Welcome to RDNets
Online high-throughput mathematical analysis of reaction-diffusion systems
News 19-10-16
Uploaded the new version 1.18 with several bug-fix and new features: fast 3D simulation with spectral method, 3D contour plot visualization, equation visualization and equation export in text format.
Follows our twitter for updates

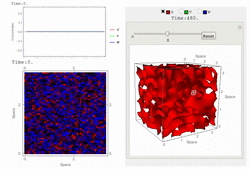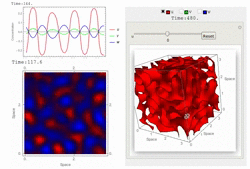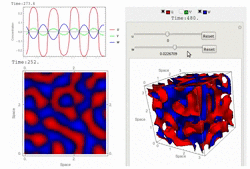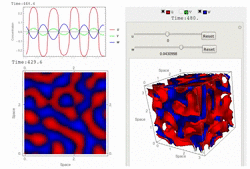
RDNets performs an automated linear stability analysis to screen for reaction-diffusion network topologies that can form self-organizing spatial patterns. The software is optimized to analyze reaction-diffusion signaling networks with cell-autonomous factors. The analysis can be constrained with qualitative and quantitative data. For more information you can read Marcon et al. 2016, PDF.
RDNets is written in Mathematica and requires only the installation of the freely available Wolfram CDF player plugin (http://www.wolfram.com/cdf-player/). The software runs on most browsers in Windows and MacOSX. Linux users need to install the Computable Data Format standalone player and run the .cdf file provided on this website.
To run the graphical user interface of RDNets, click on Start RDNets. The catalogs of possible 3-node networks and 4-node networks are also provided. The networks in these catalogs can be further constrained and analyzed with RDNets. Take a look at the video tour below for more information: